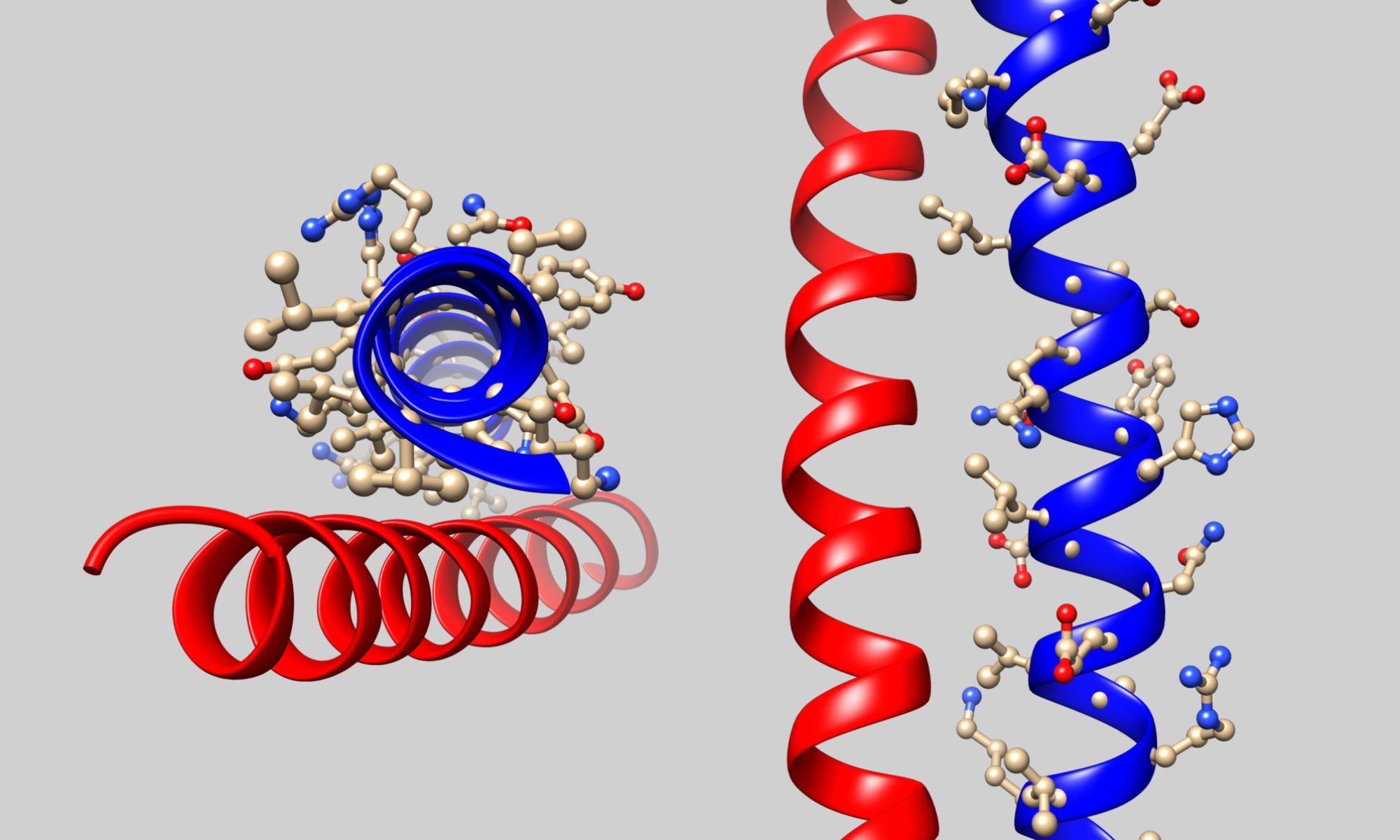
Each section title represents one of the 12 overarching themes of the Biomolecular Visualization Framework. Expand each theme for a list of the learning goals. Expand each goal for a list of the learning objectives. Please cite Dries et al. BAMBED (2017). An earlier online version of the 2017 Framework was formatted as columns.
PDF document versions: 2022, 2020, & 2017.
Atomic Geometry (AG)
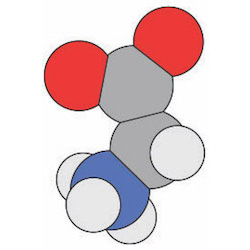
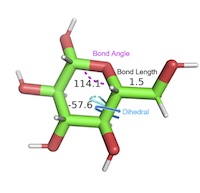 Three‐atom and four‐atom dihedral/torsion angles, metal size and metal‐ligand geometries, steric clashes. Three‐atom and four‐atom dihedral/torsion angles, metal size and metal‐ligand geometries, steric clashes. |
AG1. Students can describe the ideal geometry for a given atom within a molecule and deviations from the ideal geometry due to neighboring interactions.AG1.01 Students can identify atomic geometry/hybridization for a given atom. (Novice) AG1.02 Students can measure bond angles for a given atom. (Novice) AG1.03 Students can identify deviations from the ideal bond angles. (Amateur) AG1.04 Students can explain deviations from the ideal bond angles due to local effects. (Amateur, Expert) AG1.05 Students can predict the effect of deviations from ideal bond angles on the structure and function of a macromolecule. (Expert) AG1.06 Students can identify the geometric features of bonds (e.g. peptide bond, glycosidic, phosphoester). (Novice, Amateur) AG2. Students can compare and contrast different structural conformations with regard to energy, addition of substituents, and impact on structure/function of a macromolecule.AG2.01 Students can describe different conformations that a structure can adopt using visualization tools. (Amateur) AG2.02 Students can describe different conformations of atoms about a bond using visualization tools. (Novice) AG2.03 Students can distinguish energetically favorable and unfavorable conformations that a structure can adopt. (Amateur) AG2.04 Students can predict the effect of a given substituent on the structure and function of a macromolecule, e.g. substituent on a carbohydrate/ligand, R groups/rotamers, phosphorylation, methylation of nucleic acids, post-translational modifications. (Expert) AG3. Students can describe dihedral/torsion angles in biomolecules.AG3.01 Students can identify a dihedral/torsion angle in a three-dimensional representation of a macromolecule. (Novice) AG3.02 Students can identify the planes between which a dihedral/torsion angle exists within a three-dimensional representation of a macromolecule. (Novice) AG3.03 Students can identify phi, psi, and omega torsion/dihedral angles in a three-dimensional representation of a protein. (Amateur) |
Alternate Renderings (AR)
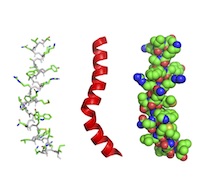 Rendering of a macromolecular structure such as a protein or nucleic acid structure in various ways from the simplest possible way (connections between alpha carbons) to illustration of secondary structure (ribbons) to surface rendering and space filling. Rendering of a macromolecular structure such as a protein or nucleic acid structure in various ways from the simplest possible way (connections between alpha carbons) to illustration of secondary structure (ribbons) to surface rendering and space filling. |
AR1. Students can interpret or create molecular images that convey features such as secondary structure, CPK coloring, and active sites.AR1.01 Students can manipulate rendered structures to illustrate molecular properties. (Novice) AR1.02 REMOVED (integrated with SF2.02) AR1.03 Students can describe or label structural differences among multiple structures. (Amateur, Expert) AR1.04 Students can infer information from rendering a structure in different ways. (Novice, Amateur, Expert) AR1.05 Students can create renderings that distinguish secondary structural features. (Novice) AR1.06 Students can create an information rich rendering of a structure that depicts structural features found in the literature. (Amateur) AR1.07 Students can create an information rich rendering of a structure containing ligands, covalent modifications, and noncanonical amino acids or nucleotides. (Amateur, Expert) AR1.08 Students can use molecular visualization to tell a story about a macromolecular structure. (Expert) AR1.09 REMOVED (integrated with MI1.02) AR1.10 Students can convert textbook images of small molecules into 3D representations in a molecular visualization program. (Amateur) AR2. Students can choose the best rendering of a macromolecule to use in a given situation.AR2.01 Students can recognize that a cartoon rendering is a summary of the detail in a line rendering. (Novice, Amateur) AR2.02 Students can describe the atoms and their representations in different renderings, e.g. coloring, showing hydrogens/double bonds. (Novice) AR2.03 Students can identify or create a suitable rendering, or combination of renderings, for a specific purpose (e.g., a surface rendering overlaid with a cartoon to highlight the van der Waals surface alongside secondary structure, or active site sticks shown over a cartoon). (Novice, Amateur) AR2.04 Students can identify the limitations in various renderings of molecular structures. (Amateur) AR2.05 Students can understand the level of detail of different molecular representations. (Novice, Amateur, Expert) AR2.06 Students can transition comfortably between equivalent 2D and 3D renderings of biomolecules. (Novice, Amateur, Expert) AR2.07 Students can use and interpret color in the context of macromolecules to clarify and/or highlight features (e.g., coloring amino acids differently by property, different molecules uniquely in a complex, protein chains, secondary structure). (Novice) |
Construction and Annotation (CA)
![[xxTODO]](https://biomolviz.org/wp-content/uploads/2020/09/CA-1gzm-thumbnail.jpg) Ability to build macromolecular models, either physical or computerized, and, where possible, add commentary, either written or verbal, to tell a molecular story. Ability to build macromolecular models, either physical or computerized, and, where possible, add commentary, either written or verbal, to tell a molecular story. |
CA1. Students can compose information‐rich renderings of macromolecule‐ligand interactions.CA1.01 Students can construct and annotate a model of a macromolecule bound to a ligand. (Amateur) CA1.02 Students can construct a model of a macromolecule bound to a ligand and identify the types of molecular interactions. (Amateur) CA1.03 Students can construct a model of a macromolecule bound to a ligand and assess the importance of molecular interactions. (Expert) CA1.04 Students can produce a model of a macromolecule based on a known structure of a related macromolecule. (Amateur, Expert) CA2. Students can compose a rendering to predict the cellular location of a protein (e.g., extracellular, membrane associated, or cytoplasmic) based on the properties and orientations of functional groups.CA2.01 Students can design a rendering that conveys properties such as polarity, charge, secondary structure, etc. to suggest the cellular location of a macromolecule. (Amateur) CA2.02 Students can create protein images with colored polar/nonpolar residues to determine whether they fold with a hydrophobic core. (Novice) CA2.03 Students can create images to display polar/nonpolar residues and propose a role for the protein and/or how it interactions with its environment ‐ and that the predictions would be plausible based on the protein. (Amateur) CA2.04 Students can make accurate predictions of the location/function of the protein that incorporates additional protein features, such as transmembrane helices, apparent docking surfaces, etc. (Expert) |
Ligands and Modifications (LM) [HG]
 Metals and metal clusters, additions such as glycosylation, phosphorylation, lipid attachment, methylation, etc. Metals and metal clusters, additions such as glycosylation, phosphorylation, lipid attachment, methylation, etc.
Note that this theme was formerly called Hetero Group Recognition (HG). |
LM1. Students can identify ligands and modified building blocks (e.g., hydroxyproline, aminosaccharides, modified nucleobase) within a rendered structure.LM1.01 Students can use the annotation associated with a pdb file to identify and locate ligands and modified building blocks in a given biomolecule. (Amateur) LM1.02 Students can visually identify non‐protein chemical components in a given rendered structure. (Amateur) LM1.03 Students can distinguish between nucleic acid and ligands (e.g. metal ions) in a given nucleic acid superstructure. (Amateur) LM1.04 Students can explain how a ligand in a given rendered structure associates with the biomolecule (i.e., covalent interaction with residue X). (Amateur) LM1.05 Students can locate/identify ligands and modified building blocks in unannotated structures and describe their role. (Expert) LM2. Students can describe the impact of a ligand or modified building block on the structure/function of a macromolecule.LM2.01 Students can look at a given rendered structure and describe how the presence of a specific ligand or modified building block alters the structure of that biomolecule. (Amateur) LM2.02 Students can explain how the removal of a particular ligand or modified building block would alter the structure of a given biomolecule. (Expert) LM2.03 Students can use molecular visualization tools to predict how a specified ligand or modified building block contributes to the function of a given protein. (Amateur) LM2.04 Students can predict how a ligand or modified building block contributes to the function of a protein for which the structure has been newly solved. (Expert) |
Macromolecular Assemblies (MA)
 Polypeptides, oligosaccharides, and nucleic acid and lipid superstructures (e.g. protein–nucleic acid complexes, lipid membrane-associated proteins). Polypeptides, oligosaccharides, and nucleic acid and lipid superstructures (e.g. protein–nucleic acid complexes, lipid membrane-associated proteins). |
MA1. Students can describe various macromolecular assemblies.MA1.01 Students can identify individual biomolecules in a macromolecular assembly. (Novice, Amateur, Expert) MA1.02 Students can describe functions of individual biomolecules within a macromolecular assembly. (Novice, Amateur, Expert) MA1.03 Students recognize the various lipid ultrastructures (e.g. micelles, bicelles, vesicles, and lipid bilayers) in a 3D structure. (Novice) MA2. Students can compose information‐rich renderings of macromolecular assemblies.MA2.01 Students can render a macromolecular assembly to highlight individual structures. (Amateur) MA2.02 Students can render a macromolecular assembly to illustrate structural features, (e.g. binding interfaces, symmetry, tertiary structure, etc). (Amateur, Expert) |
Macromolecular Building Blocks (MB) [MR]
 Recognition of native amino acids, nucleotides, sugars, and other biomonomer units/building blocks. Understanding of their physical and chemical properties, particularly regarding functional groups. Recognition of native amino acids, nucleotides, sugars, and other biomonomer units/building blocks. Understanding of their physical and chemical properties, particularly regarding functional groups.Note that this theme was formerly called Monomer Recognition (MR). |
MB1. Students can identify individual building blocks of biological polymers.MB1.01 Given a rendered structure of a biological polymer, students will be able to identify the ends of a biological polymer. (Novice, Amateur, Expert) MB1.02 Given a rendered structure, students can divide the polymer into its individual building blocks. (Novice) MB1.03 Given a rendered structure, students can identify the individual building blocks. (Novice) MB2. Students can describe the contributions different individual building blocks make in determining the 3D shape of the polymer.MB2.01 Students can describe the physical/chemical properties of an individual building block/functional group in a rendered structure of a polymer. (Amateur) MB2.02 Students can describe the significance of the location of individual building blocks within the 3D structure of a polymer (e.g. protein, carbohydrate or nucleic acid). (Novice, Amateur, Expert) MB2.03 Students can identify physical/chemical properties of individual building blocks/functional groups in different local environments. (Amateur) MB2.04 Using a visualized structure, students can identify stereochemistry (e.g. in carbohydrate, lipid, and protein structures). (Amateur) MB2.05 Students can modify/mutate a building block to change the 3D structure of a polymer (e.g. protein, carbohydrate or nucleic acid). (Amateur, Expert) |
Molecular Dynamics (MD)
 Animated motion simulating conformational changes involved in ligand binding or catalysis, or other molecular motion/dynamics. Animated motion simulating conformational changes involved in ligand binding or catalysis, or other molecular motion/dynamics. |
MD1. Students can describe the impact of the dynamic motion of a biomolecule on its function.MD1.01 Students can recognize that biological molecules have different conformations. (Novice, Amateur) MD1.02 Students can correlate molecular movement with function. (Novice, Amateur, Expert) MD2. Students can predict limits to macromolecular movement.MD2.01 Students can locate potential regions of flexibility and inflexibility in the structure of a biomolecule. (Novice, Amateur) MD2.02 Students can recognize acceptable/unacceptable movement within a macromolecule by determining whether the movement is within allowable bond angles. (Expert) MD2.03 Students can recognize acceptable/unacceptable movement within a macromolecule by determining whether the movement results in steric hindrance. (Amateur) MD2.04 Students can recognize acceptable/unacceptable movement within a macromolecule by considering the atomic packing constraints. (Expert) |
Molecular Interactions (MI)
 Covalent and noncovalent bonding governing ligand binding and subunit‐subunit interactions. Covalent and noncovalent bonding governing ligand binding and subunit‐subunit interactions. |
MI1. Students can predict the existence of an interaction using structural and environmental information (e.g. bond lengths, charges, pH, dielectric constant)MI1.01 Students can distinguish between covalent and noncovalent interactions. (Novice) MI1.02 Students can identify the different non‐covalent interactions (e.g., hydrogen bonds, ionic interactions, van der Waals contacts, induced dipole) given a 3D structure. (Amateur) MI1.03 Students can predict whether a functional group (region) would be a hydrogen bond donor or acceptor. (Amateur) MI1.04 Students can render the 3D structure of a biomolecule so as to demonstrate the ionic interactions and/or charge distribution of the different non‐covalent interactions. (Amateur) MI1.05 As it relates to a particular rendered structure, students can rank the relative strengths of covalent and noncovalent interactions. (Amateur) MI2.Students can evaluate the effect of the local environment on various molecular interactions.MI2.01 Students can identify regions of a biomolecule that are exposed to or shielded from solvent. (Novice) MI2.02 Students can identify other molecules in the local environment (e.g. solvent, salt ions, metals, detergents, other small molecules) that impact a molecular interaction of interest. (Novice) MI2.03 Students can predict the impact of other molecules in the local environment (e.g. solvent, salt ions, metals, detergents, other small molecules) on a molecular interaction of interest. (Amateur) MI2.04 Students can predict the pKa of an ionizable group based on the influence of its local three-dimensional environment. (Amateur) MI2.05 Students can propose a change to the local environment that would yield a desired change in a molecular interaction. (Expert) MI2.06 Using molecular visualization tools, students can determine which intermolecular force is most critical to stabilizing a given interaction. (Expert) |
Symmetry/Asymmetry Recognition (SA)
![[xxTODO]](https://biomolviz.org/wp-content/uploads/2020/09/SA-6sl0-thumbnail.jpg) Recognition of symmetry elements within both single chain and oligomeric multi-chain macromolecules. Recognition of symmetry elements within both single chain and oligomeric multi-chain macromolecules. |
SA1. Students can identify symmetric or asymmetric features in rendered molecules.SA1.01 Students can identify symmetric features in a rendered molecule (shown in fixed orientation). (Novice) SA1.02 Students can rotate a single macromolecule, multi-chain macromolecules (e.g., homo- or heteromers), complexes of macromolecules, and supramolecular assemblies to identify axes of symmetry. (Amateur) SA1.03 Students can identify symmetric and asymmetric features in rendered molecules after coloring a given rendered molecule to reveal structural features (charge, hydrophobicity, etc.). (Amateur) SA2. Students can hypothesize the functional significance of symmetry or asymmetry in rendered molecules.SA2.01 Students can explain the functional significance of rotational axes of symmetry (or asymmetry) in a given rendered molecule. (Novice, Amateur, Expert) SA2.02 Students can predict the functional significance of symmetry (or asymmetry) in a given rendered molecule. (Amateur, Expert) |
Structure‐Function Relationship (SF)
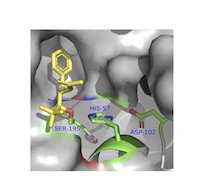 Active/binding sites, microenvironments, nucleophiles, redox centers, etc. Active/binding sites, microenvironments, nucleophiles, redox centers, etc.Note: please also see LM2.03. |
SF1. Students can evaluate biomolecular interaction sites using molecular visualization tools.SF1.01 Students can identify functionally relevant cofactors, ligands or substrates associated with a macromolecule and describe their role (e.g., an active site magnesium ion). (Amateur, Expert) SF1.02 Students recognize that the size and shape of the ligand must match the size and shape of the binding site. (Novice, Amateur) SF1.03 Students recognize that the polarity or electrostatic potential of a surface complements that of the ligand or substrate. (Novice, Amateur) SF1.04 Students recognize that the hydrophobicity of a surface complements that of the ligand or substrate. (Novice) SF1.05 REMOVED (integrated with SF1.03). SF1.06 Students can use docking software to predict how the surface properties of a macromolecule guides and allows the binding of a ligand or substrate. (Amateur) SF2. Using molecular visualization, students can predict the function of biomolecules.SF2.01 Students can recognize structurally related molecules. (Novice) SF2.02 Students can superimpose structurally related molecules. (Novice, Amateur) SF2.03 Students can identify functionally relevant features of a macromolecule (e.g., an active site cysteine, a functional loop) (Amateur) SF2.04 Students can predict molecular function given a binding site. (Amateur, Expert) SF3. Using molecular visualization, students can predict the function of an altered macromolecule.SF3.01 Students can structurally alter a macromolecule. (Novice) SF3.02 Students can propose structural alterations to test interactions in a macromolecule. (Amateur) SF3.03 Students can predict the impact of a structural alteration on the function of a macromolecule. (Amateur, Expert) |
Structural Model Skepticism (SK)
![[xxTODO]](https://biomolviz.org/wp-content/uploads/2020/09/SK-1fcc-thumbnail-300x284.jpg) Recognition of the limitations of models to describe the structure of macromolecules. Recognition of the limitations of models to describe the structure of macromolecules. |
SK1. Students can critique the limitations of a structural model of a macromolecule.SK1.01 Students can explain that the pdb file is a model based on data and that, as a model, it has limitations. (Novice, Amateur) SK1.02 Students associate resolution with reliability of atom positions. (Amateur) SK1.03 Students can identify building blocks (for example, amino acid sidechains) whose orientation in a biopolymer is uncertain. (Expert) SK1.04 Students can evaluate the flexibility/disorder of various regions of a macromolecular structure. (Novice, Amateur, Expert) SK1.05 Students can reconcile inconsistent numbering of individual building blocks among species and structure files. (Novice) SK1.06 Students can utilize a Ramachandran plot/steric clashes to interpret the validity of a structure. (Amateur, Expert) SK1.07 Students can describe the limitations of a macromolecule‐ligand docking simulation. (Amateur, Expert) SK2. Students can evaluate the quality of 3D models including features that are open to alternate interpretations based on molecular visualization and PDB flat files.SK2.01 Students can evaluate a crystal structure for crystal packing effects. (Novice, Amateur, Expert) SK2.02 Students can resolve differences between the asymmetric unit and the functional biological assembly. (Expert) SK2.03 Students can differentiate functional ligands (with biological/biochemistry role) from nonfunctional ligands (e.g. most solvents, salts, ions, and crystallization agents). (Novice, Amateur, Expert) SK3. Students can discuss the value of experimentally altering a biomolecule to facilitate structure determination.SK3.01 Students can identify non‐native structural features. (Amateur) SK3.02 Students can propose molecular modifications to facilitate structure determination. (Amateur, Expert) SK3.03 Students can propose a purpose for the introduction of non‐native structural features to facilitate structure determination. (Amateur, Expert) |
Topology and Connectivity (TC)
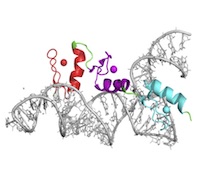 Following the chain direction through the molecule, translating between 2D topology mapping and 3D rendering. Following the chain direction through the molecule, translating between 2D topology mapping and 3D rendering. |
TC1. Students can describe the linkages between building blocks within a macromolecule.TC1.01 Students can trace the backbone of a macromolecule in three dimensions. (Novice, Amateur) TC1.02 Students can use appropriate terms to describe the linkages/bonds/interactions that join individual building blocks together in a macromolecule or macromolecular assembly. (Novice, Amateur) TC1.03 Given a virtual model of individual building blocks, students can predict the types of linkages/bonds/interactions that are possible or favorable. (Amateur) TC1.04 Given individual building blocks, students can appropriately connect them to create a biological polymer (e.g., drawing carbohydrate linkages, a small peptide). (Amateur, Expert) TC2. Students can describe the overall shape and common motifs within a 3D macromolecular structure.TC2.01 Using molecular visualization software, students can describe the three-dimensional structure of a macromolecule, including overall shape and common structural motifs. (Novice, Amateur, Expert) TC2.02 Students can identify common domains/motifs within a macromolecule. (Amateur, Expert) TC2.03 Students can identify connectivity features between domains or subunits in a macromolecular structure. (Amateur) TC2.04 Students can identify interactions between domains or subunits in a macromolecular structure. (Amateur, Expert) TC2.05 Students can describe how domains/motifs in a macromolecule work together to achieve a concerted function in the cell. (Amateur, Expert) TC2.06 Students can identify the levels of protein structure (e.g., parse a tertiary/quaternary structure into a series of secondary structures/motifs) and the ways in which they are connected from a three‐dimensional structure. (Novice, Amateur, Expert) TC3. Students can explain how any given biomolecular interaction site can be made by a variety of topologies.TC3.01 Students can recognize that the groups that comprise a functional site only require proper arrangement in three dimensional space rather than a particular order or position in the linear sequence. (Amateur) TC3.02 Students can recognize similarities and differences in two similar ‐ but not identical ‐ three dimensional structures. (Amateur) TC3.03 Students can describe dissimilar portions of homologous proteins as arising from genetic insertions/deletions/rearrangements. (Amateur) |
Last updated 2-Jun-2022