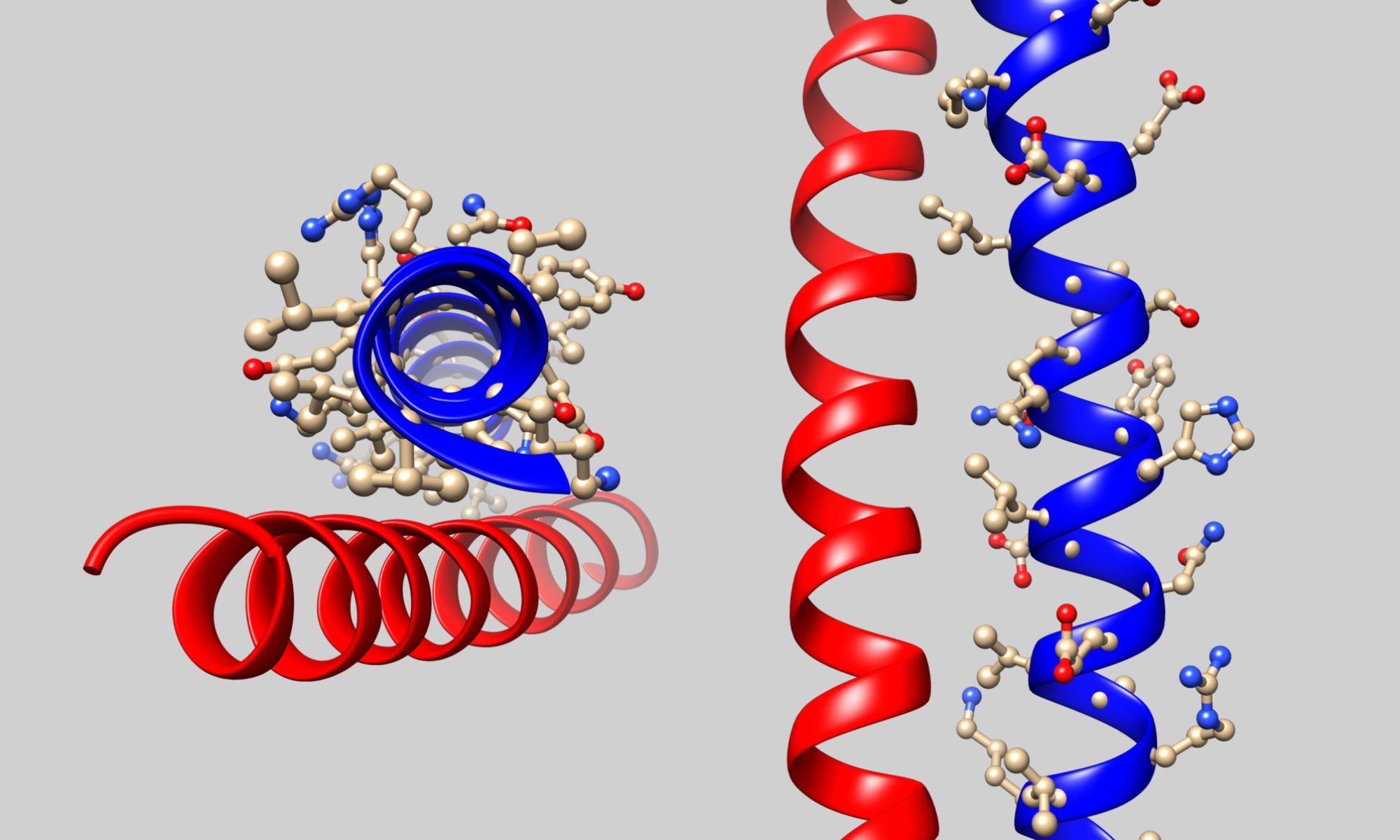
If you are a biochemistry educator looking for resources to teach online, we recommend the Association for Biochemistry and Molecular Biology (ASBMB) “Online Teaching: Practices and Resources” website, co-authored by some BioMolViz team members. This site contains links to biomolecular visualization tools, and other tools for teaching biochemistry online.
Online Biomolecular Visualization Tools
The following tools allow the user to find and create models without learning a great deal of code or a new visualization program. These resources are highly recommended for instructors.
Proteopedia (e.g. DNA ligase): The Proteopedia Wiki is an encyclopedia of macromolecules that allows easy interaction with 3D models. The scene-authoring tool allows users to create interactive scenes without having to download a modeling program or learn a new script.
Visit the BioMolViz Proteopedia page where we are curating Framework theme examples from Proteopedia entries.
Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB) Molecule of the Month (e.g. ATP synthase): “The RCSB PDB Molecule of the Month by David S. Goodsell (RCSB PDB-Rutgers and The Scripps Research Institute) presents short accounts on selected molecules from the Protein Data Bank. Each installment includes an introduction to the structure and function of the molecule, a discussion of the relevance of the molecule to human health and welfare, and suggestions for how visitors might view these structures and access further details.”
RCSB PDB Ligand Explorer 3D (e.g. Human PPAR alpha ligand binding domain): Most, if not all, PDB entries allow you to access the “Ligand Explorer” from the main entry page. This built-in web tool allows for quick 3D visualization. The user can zoom in and out, rotate the model, and display macromolecule-ligand interactions with a click.
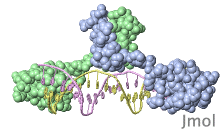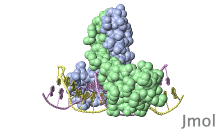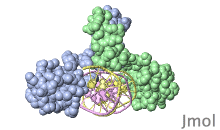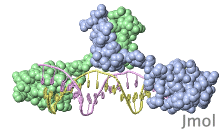
FirstGlance.Jmol.Org is an informative 3D visualization tool for beginners to explore any protein structure—yet it is powerful enough for researchers. Minimizing (and explaining) technical terminology, it lets the user concentrate on understanding protein structure rather than on how to use the software. In order to be as easy as possible to use, FirstGlance in Jmol does not provide tools to customize the molecular view, but rather offers a series of “canned” views that reveal the major structural features of the molecule. Any molecular scene obtained in FirstGlance in Jmol can be saved, with just a few clicks, as a high-resolution static image, or as a presentation-ready animation.
iCn3D: Similar to the ligand explorer above, iCn3D (“I can see in 3D”) is a WebGL-based viewer for interactive viewing of 3D macromolecular structure without the need to install a separate application. With this software you can interactively view 3D structures and corresponding sequence data, interactively view 3D alignments of similar structures, customize the display of a structure, and generate a URL that allows you to share the link and incorporate iCn3D into your own pages.
CellPAINT allows the user to create dynamic illustrations of cells and viruses. This resource was developed for an initiative to model whole living cells.

The Online Macromolecular Museum: The Online Macromolecular Museum (OMM) is a site for the display and study of macromolecules. Macromolecular structures, as discovered by crystallographic, NMR, or Cryo-EM methods, are scientific objects in much the same sense as fossil bones or dried specimens: they can be archived, studied, and displayed in aesthetically pleasing, educational exhibits. Hence, a museum seems an appropriate designation for the collection of displays that we are assembling. The OMM’s exhibits are interactive tutorials on individual molecules in which hypertextual explanations of important biochemical features are linked to illustrative renderings of the molecule at hand.
Guides for Program-Based Molecular Visualization Tools
BioMolViz YouTube Channel
Subscribe to BioMolViz’s YouTube channel, where we feature tools for macromolecular modeling and assessment of visual literacy.
PyMOL
This Guide to creating rotatable images in PyMOL is for integrating images into Microsoft Word and PDF documents. This is ideal for providing students with a digital 3D image in a handout.
PyMOL selections tutorial video: Intro to the PyMOL GUI (14:23 min) – An instructional video on how to use the PyMOL graphical user interface (GUI). This shows the user what they can create with just a mouse with almost no command line necessary.
Essential External PyMOL Pages: The PyMOL Wiki ~ PyMOL Advanced Scripting Workshop by Schrödinger
Jmol/JSmol
CREST Jmol Tutorials Resources: a collection of digital and written guides, accompanied by video tutorials to help the user learn JMol basics.
Essential External Jmol Pages: The Online Macromolecular Museum Jmol Scripting Exhibit ~ Jmol/JSmol Interactive Scripting Documentation (St. Olaf)
iCn3D (“I see in 3D”)
Essential External iCn3D Page: NCBI iCn3D help page
UCSF ChimeraX
Essential External ChimeraX Pages: UCSF ChimeraX User Guide ~ ChimeraX Tutorials
Helpful Resources for Image/Assessment Creation
Colorblindness: If you are not colorblind, it is difficult to image how your images appear to someone with colorblindness. BioMolViz uses the Coblis—Colorblindness Simulator to inspect our images for colorblind-friendliness. Examining images using the deuteranomaly filter—the most common type of red-green colorblindness—is a great starting point for the instructor seeking to make their colored images more equitable.